Capture chromatin topology anchored at gene promoter sites
Pan Promoter Kit
What is Targeted Promoter Enrichment?
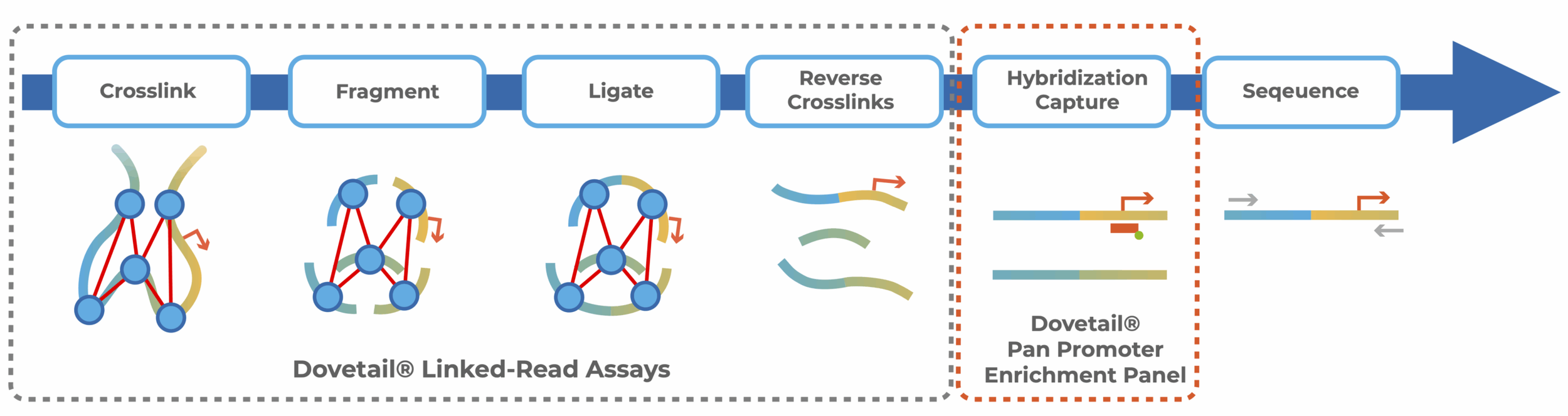
Next-generation sequencing (NGS) provides powerful, genome-wide insights. But sometimes, precision matters most. Targeted enrichment enables focused investigations by hybridizing sequencing libraries to capture probes targeting pre-selected genomic regions. These captured duplexes are then pulled down prior to sequencing, enriching your library and reducing the sequencing required to achieve meaningful coverage of your regions of interest.
Enhanced by Linked-Read libraries
When combined with Linked-Read technologies, targeted promoter enrichment becomes even more powerful. Traditionally, this has been done using Hi-C. However, Hi-C introduces a challenge: unevenly distributed restriction sites that create coverage bias, complicating enrichment panel design and data interpretation.
Dovetail® Linked-Read libraries eliminate this problem. By using sequence-independent approaches, our technology removes restriction site bias, enabling you to apply standard panel design principles and achieve consistent, accurate coverage across target regions.
The Dovetail® Pan Promoter Kit is compatible with Dovetail® LinkPrep™ Kit and Dovetail® Micro-C Kit.
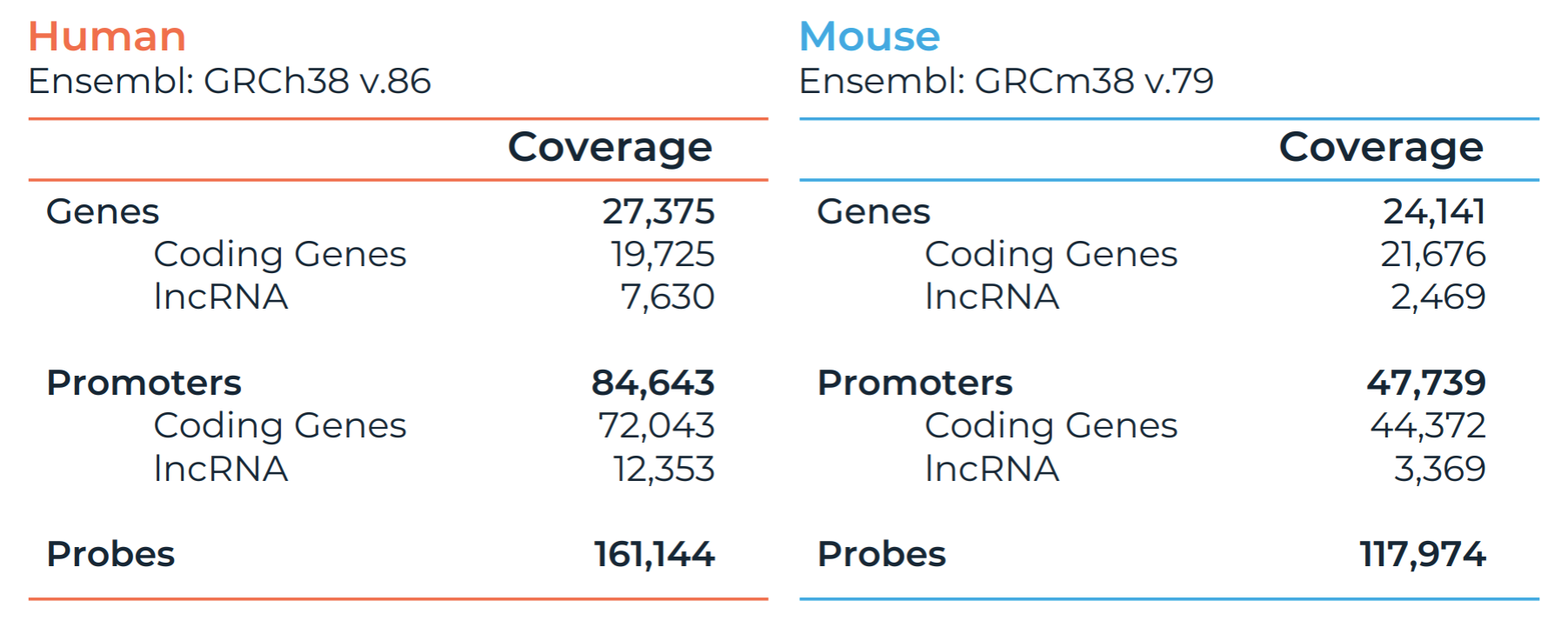
Comprehensive Panel Coverage
Pan Promoter Panels are optimized to capture all promoter regions across human or mouse genomes. This enables researchers to directly link data from ChIP-seq, ATAC-seq, or GWAS to their corresponding promoters—accelerating discovery and simplifying interpretation.
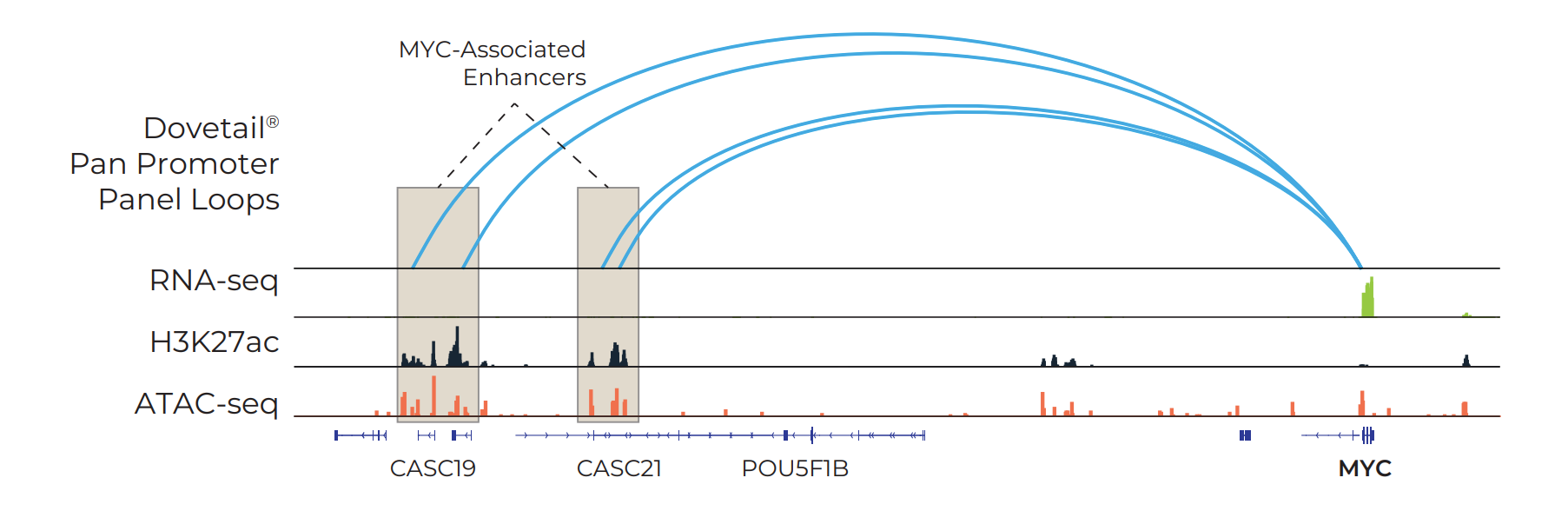
View Gene Expression in a New Dimension
NGS data types such as ChIP-seq and ATAC-seq are blind to the 3D architectural structures that connect promoters and enhancers across long genomic distances.
The power of promoter-targeted linked-read data is illustrated in the figure: interactions anchored at the MYC proto-oncogene promoter span more than 700 kb of sequence, identifying upstream regulatory regions. These regions align with enhancer (H3K27ac) and open chromatin (ATAC-seq) marks, while enhancer marks located closer to the MYC gene do not exhibit evidence of interaction.
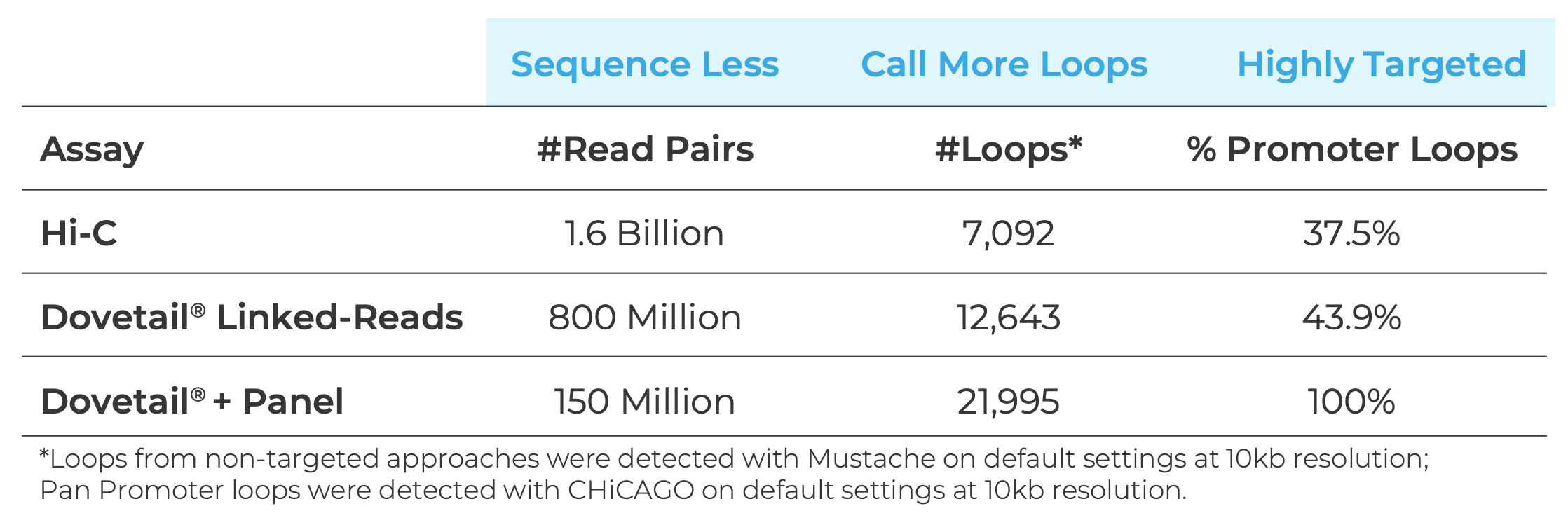
Sequence Less and Discover More, While Simplifying Data Analysis
Whole-genome experiments can require well over 1 billion read pairs to detect chromatin interactions. As a result, many studies are either underpowered or prohibitively expensive. By anchoring sequenced reads to promoters, sequencing depth requirements are reduced tenfold compared to non-targeted approaches, and the resulting data is more sensitive.
Compared to whole-genome data, a greater number of interactions are detected, with virtually all data being informative of promoter regions. Data generated using the Pan Promoter Panel integrates easily with open-source tools, enabling simple and robust analysis.
Dr. Orlando Contreras-López – Research Scientist at NGI Stockholm – SciLifeLab
"The quality of service from the Dovetail® scientific support team is one of the best we have experienced from vendors. They show a great deal of technical knowledge and a genuine care in supporting our efforts to prepare high quality proximity libraries using Dovetail® kits."
Webinar Highlight
Enriching our understanding of gene regulation—a targeted view linking promoters with their regulatory elements.
The Dovetail® Pan Promoter Panels can be used when a targeted approach is desired because the hypothesis is more narrowly defined, reducing overall study costs.
Often referred to as promoter capture Hi-C, this method enriches for genomic regions of interest by ‘pulling down’ pre-selected genomic regions with capture probes before next-generation sequencing (NGS).
Contact us.
email@example.com
(555) 555-5555
123 Demo Street
New York, NY 12345