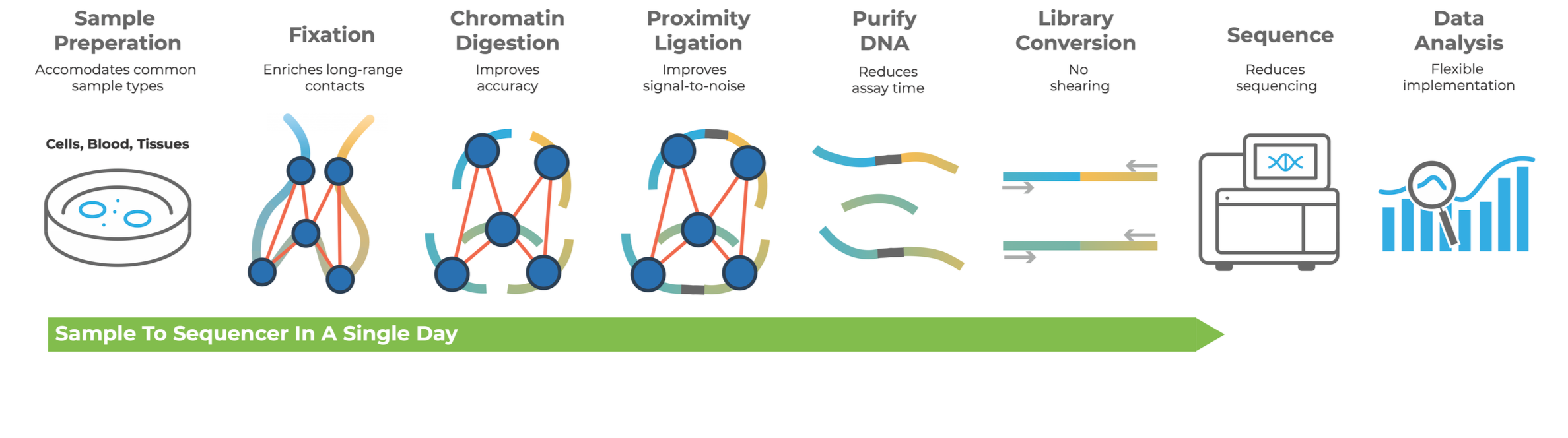
Meet LinkPrep Technology—an all in one linked-read assay bringing you fast, highly sensitive results, with less bias and more resolution than traditional Hi-C methods. Capture long-read information, with short-read sequencing. Empower your research with our complete solution.
Meet the Dovetail® LinkPrep™ Kit
An Integrated Solution
Dovetail® LinkPrep™ technology expands the capabilities of short-read sequencers to capture long-range genetic information. It enables base pair-resolved genetic variation detection, high-resolution 3D genome characterization, and rapid haplotype-aware assemblies. Utilizing standard NGS workflows, the LinkPrep™ Assay generates linked-read data, enhancing sensitivity and improving accuracy across a broad range of applications to advance your research.
Simple, fast workflow
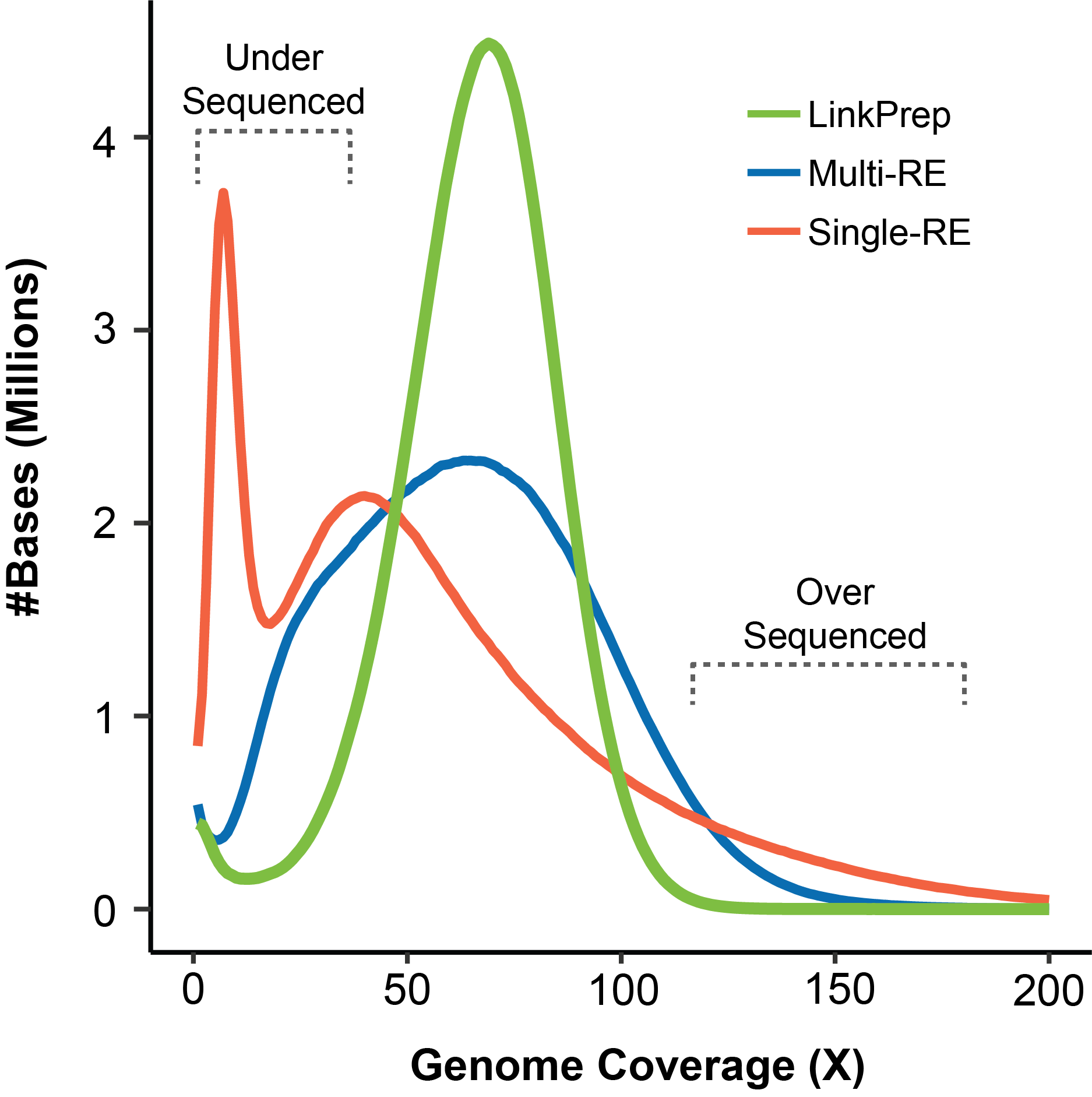
LinkPrep™ technology delivers improved genome coverage and data quality compared to traditional short-read sequencing approaches. The enhanced signal-to-noise ratio (green curve) underscores the assay’s ability to achieve higher resolution and more accurate detection of structural variations and chromatin interactions. This capability offers a clearer understanding of complex genomic regions, enabling you to extract more meaningful insights from your data.
Designed for simplicity and speed, the Dovetail® LinkPrep™ Kit includes all the critical components needed to produce sequencer-compatible libraries in a single-day workflow.
Explore Applications
Genome Assembly
Epigenetics
Genetic Variation
Genetic Variation
Improved Whole Genome Variant Discovery with Dovetail® LinkPrep™ Technology
How does LinkPrep support somatic variant detection?
Dovetail® LinkPrep™ technology enhances somatic variant detection by providing a more sensitive and comprehensive view of the cancer genome. Traditional short-read sequencing often struggles to capture complex structural changes or low-frequency variants present in heterogeneous tumor samples. LinkPrep overcomes this limitation by linking short reads into long-range information, enabling base-pair–resolved detection across the full spectrum of variants—including SNVs, InDels, CNVs, and SVs—in a single assay.
With this approach, researchers can:
Capture all relevant variants: Detect subtle point mutations alongside large, complex rearrangements that drive tumor progression.
Increase sensitivity for rare events: Linked-read context improves resolution for low-abundance somatic mutations within mixed cell populations.
Retain WGS-like capabilities: Deliver standard coverage for SNVs, InDels, and CNVs while enabling ultra-high sensitivity for structural variants often missed in conventional sequencing.
Simplify workflows: Generate data through standard NGS pipelines—no specialized equipment or training required—making high-resolution variant detection accessible to any lab.
By delivering both breadth and accuracy, LinkPrep™ gives cancer researchers an unparalleled ability to refine driver mutation lists, build more complete variant catalogs, and advance insights into tumor biology and therapeutic targeting.
LinkPrep for Genetic Variation is a complete solution - from sample processing to bioinformatic insights.
Customer Testimonial
"Our lab is excited about the potential of LinkPrep™ data. We currently use a range of NGS-based assays to catalog genetic variations spanning from SNVs up to large SVs. LinkPrep™ has the potential to become a key tool in our oncology research, offering powerful capabilities for the de novo detection of the full spectrum of somatic variants that we focus on."
Brian Walker, Ph.D.
Professor of Medical and Molecular Genetics at Indiana University School of Medicine
Explore our oncology customer highlight
Epigenetics
Revolutionize your understanding of gene regulation and biological processes through the power of 3-dimensional DNA organization.
What LinkPrep Provides for 3D genomics?
LinkPrep data quality means you detect more features (TADs and loops) compared to traditional Hi-C, even at reduced sequencing depths. As a result, fewer sequencing libraries and less sequencing is required simplifying experimental design and execution.
Produce highly uniform sequence coverage across all parts of the genome. Where the non-uniform restriction enzyme site distribution associated with traditional Hi-C methods often skews signals, you can be confident in the position of interactions detected by LinkPrep.
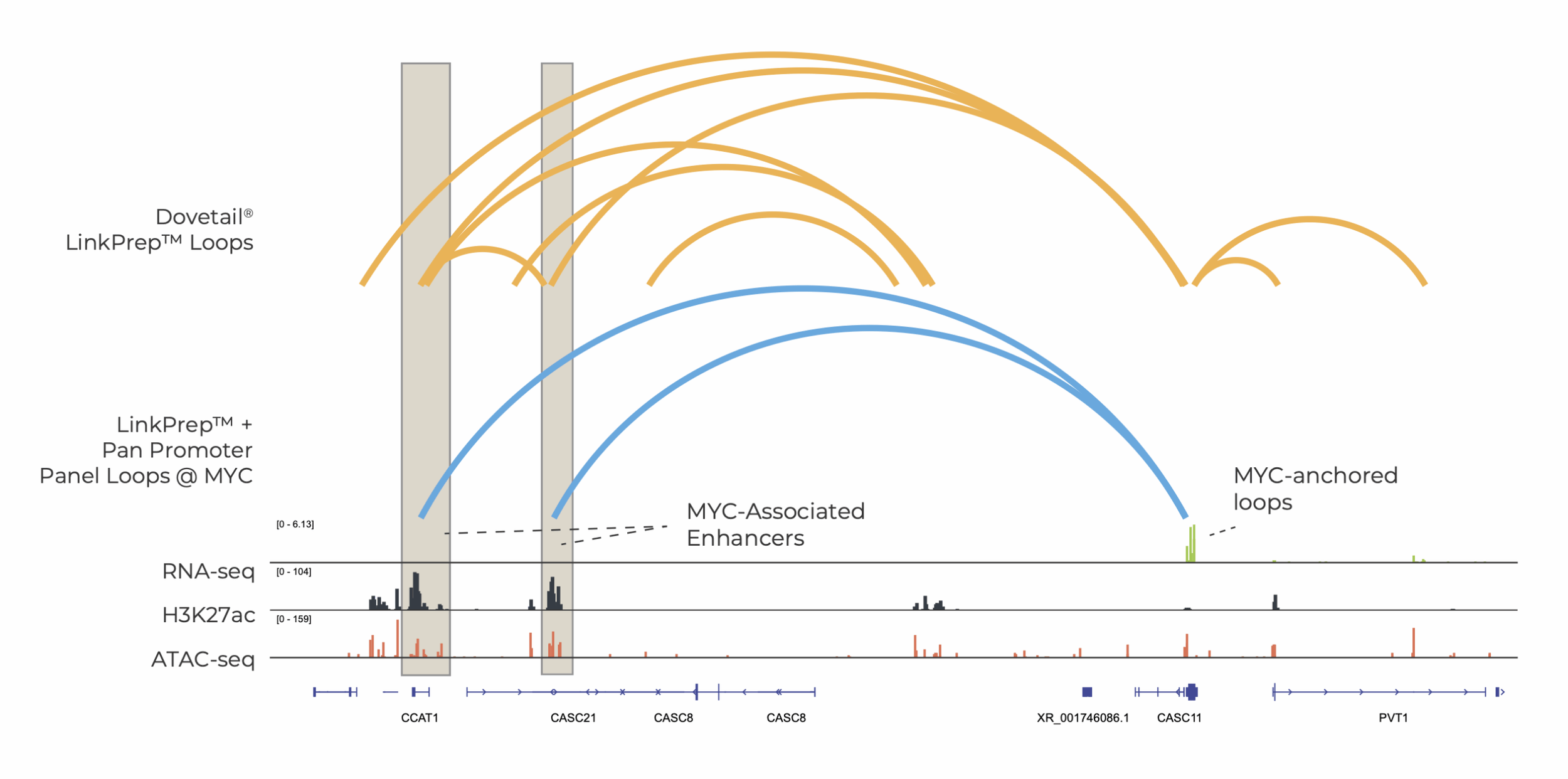
LinkPrep is now compatible with hybrid capture
The Dovetail® LinkPrep™ Assay now supports hybrid capture. When paired with the Dovetail® Pan Promoter Panel, targeted LinkPrep™ data enables efficient promoter-anchored interaction profiling and enriches biologically meaningful signals, revealing connections between promoters, regulatory elements, and non-coding variants.
This powerful approach can accelerate research in GWAS, oncology, and 3D genomics. Whether you're using our Pan Promoter Panel, an off-the-shelf third-party panel, or your own custom design, targeting LinkPrep interactions delivers high-quality data, streamlined analysis, and scalability—all while maintaining the speed, accuracy, and affordability of the core assay.
Customer Testimonial
“I have found the Dovetail® LinkPrep™ Kit to be easy to use and not affected by tissue or cell morphology. It’s been very well suited to my study of looping interactions compared to the other kits I have used.”
Benjamin Lebeau
Research Assistant, Nanyang Technological University
Genome Assembly
Reduce Time and Improve Genome Assembly Quality with the Dovetail®AssemblyLink™ Assay
How does LinkPrep enable genome assembly?
The Dovetail LinkPrep™ Assay streamlines the genome assembly process by rapidly generating Hi-C-like libraries essential for genome scaffolding. All users with standard molecular biology training can access the assay due to the simplicity of the workflow. The kit is configured to support all steps from sample to sequencing library.
Rapid workflow – sample to library in one day.
Simple to use – accessible to novice users.
Low sample input – requires as little as 5 mg of tissue.
No sonication required - specialized equipment not necessary.
Automation compatible – amenable to high-throughput settings.
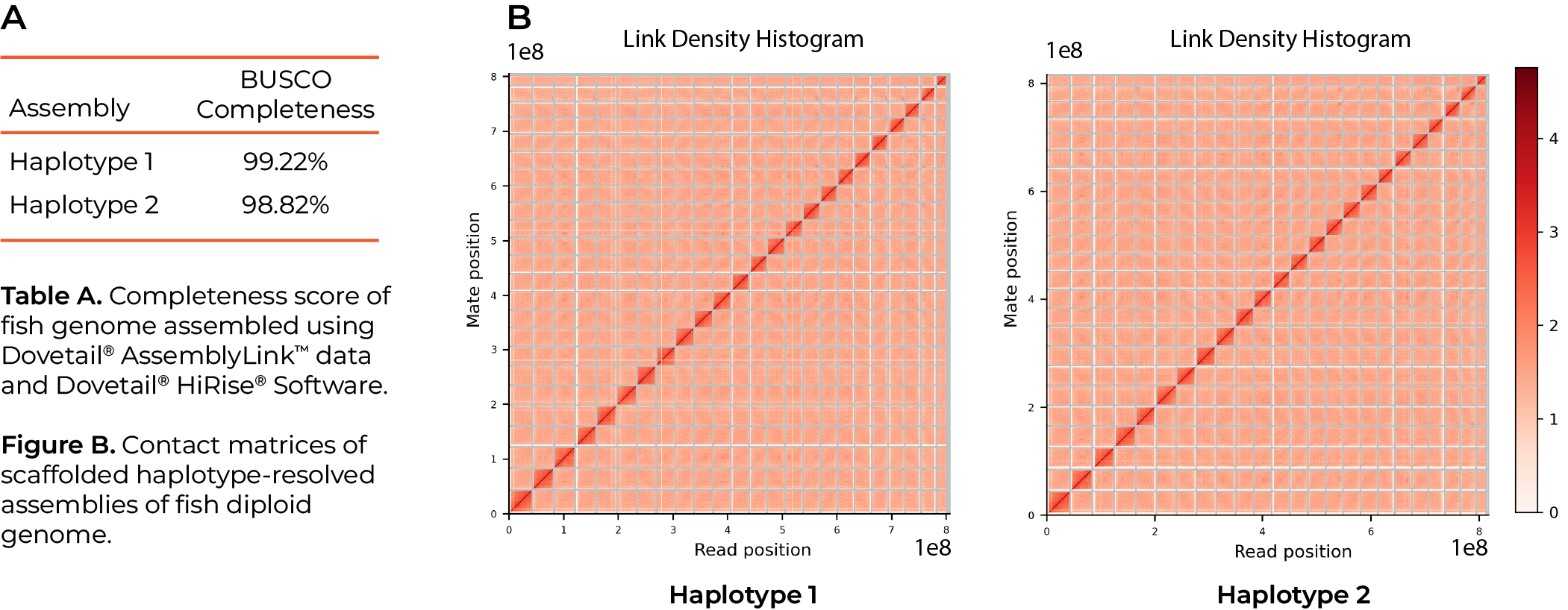
Produce Haplotype Resolved Assemblies
The LinkPrep kit offers a faster workflow that does not compromise on data quality. The assay delivers highly uniform sequence coverage facilitating variant phasing critical to producing true haplotype resolved assemblies. Moreover, this unique attribute leads to an easier path to chromosome-scale assembly of higher ploidy genomes.
High quality data – supports chromosome scale assembly.
Capture phase information – enables haplotype resolved and polyploid genome assembly.
Uniform sequence coverage – lowers sequencing depth requirements.
Customer Testimonial
"We're thrilled to onboard the Dovetail LinkPrep™ kit. The Minderoo OceanOmics Centre at UWA is using Dovetail Hi-C preps to scaffold whole genome assemblies of marine vertebrate species, as we create a comprehensive library of high-quality reference genomes. The refined workflow will decrease hands-on time in the lab and increase sequencing throughput. The reduced input requirements from fresh frozen animal tissue means we can now tackle even the smallest specimens without compromising sequencing quality or complexity."
Adrianne Doran,
Scientific Officer, Minderoo OceanOmics Centre at UWA
Customer Testimonial
“The EU-funded project Biodiversity Genomics Europe aims to create a decentralized infrastructure in Europe with a crucial component of reference genome sequencing that aims to generate around 500 genomes representing European eukaryotic biodiversity. Having a workflow that scales will be important to achieving this goal. We are excited about the speed and quality of data promised by the new Dovetail LinkPrep for Assembly Assay and look forward to the opportunity to apply it within the project.”
Ave Tooming-Klunderud
Lab and Project manager at Norwegian Sequencing Centre and Biodiversity Genomics Europe participant