Omni-C Kit
700+ Genome Assemblies
Produce the highest quality
genome assembly.
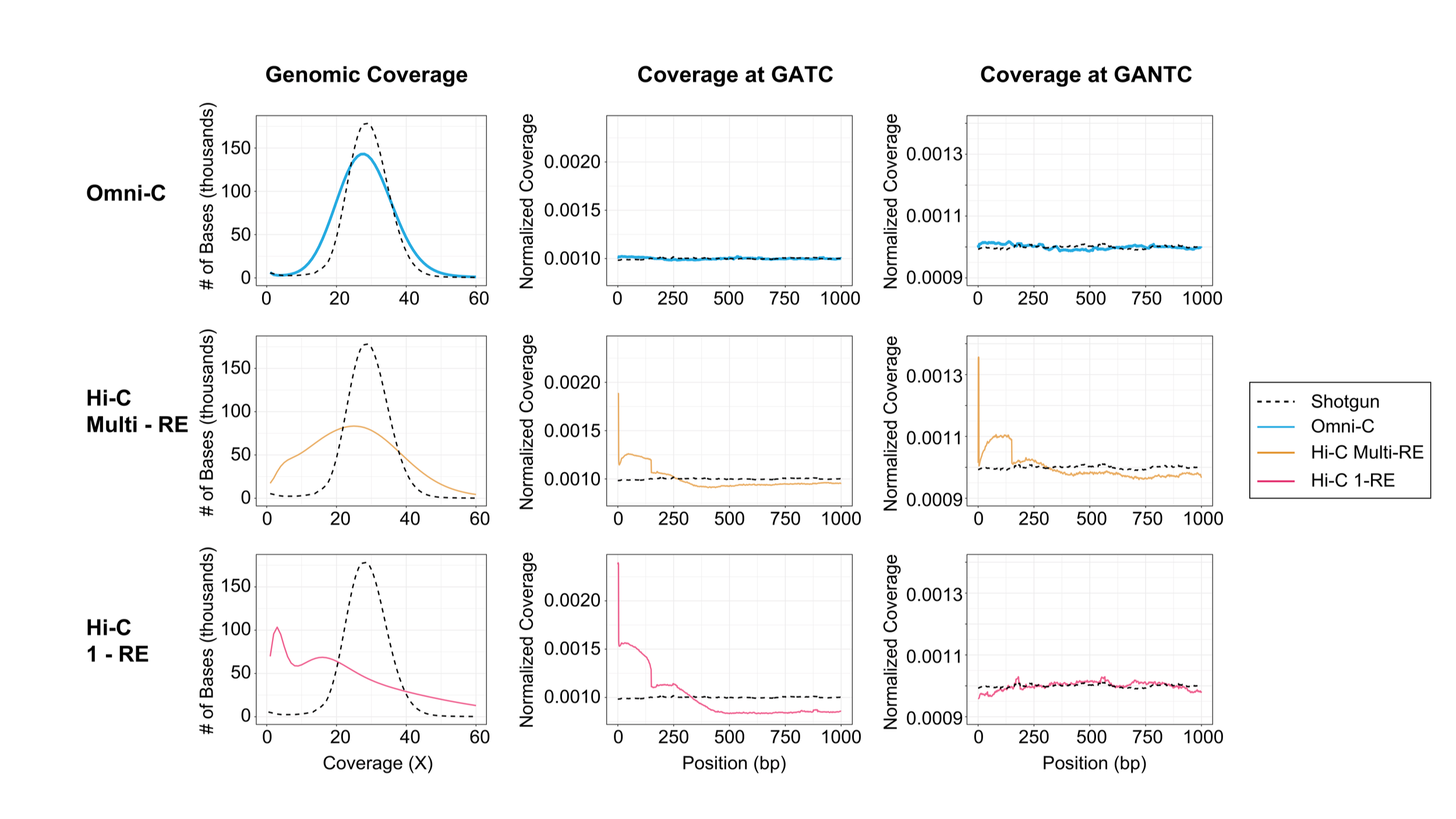
For those looking for the ultimate scaffolding tool, Dovetail Omni-C data delivers uniform, shotgun-like sequence coverage enabling genotyping and haplotype phasing, alongside long-range information characteristic of Hi-C data. Take your assembly to the next level with a fully haplotype-resolved genome.
The Omni-C kit has optimized DNase I, a sequence-independent endonuclease, to reproducibly digest chromatin, delivering all the characteristics of a Hi-C approach without the sequence bias inherent to restriction enzyme-based Hi-C approaches.
Get enriched long-range cis reads, and more complete contact matrices when viewing chromatin conformation and looping interactions. Omni-C data gives you the most complete genome-wide view for all your genome assembly, phasing, structural variant, and SNP research.
Chromosome scale haplotype assemblies of genomes with HiFi and Omni-C technologies
Haplotype-resolved assembly provides a complete picture of genomes and genetic variations. Most of the existing assembly algorithms either provide haplotype collapsed assembly or require pedigree information for phasing. New long-range sequencing approaches, such as PacBio’s HiFi technology , combined with Dovetail Omni-C libraries, a proximity ligation technology that captures chromosome-scale interactions with uniform sequencing coverage comparable to traditional shotgun sequencing libraries, make full chromosome scale haplotype resolved genome assemblies possible.
With these emerging datatypes coupled with a computational pipeline, we generated chromosome-scale phased assembly for NA12878 haplotypes with ~ 148.5 Mbp scaffold N50 for each haplotype assembly. Our assemblies phased ~ 92% of total heterozygous SNVs with more than 99% accuracy on each chromosome and showed high base-level agreement with the reference genome. Similar results were obtained for another human genome sample (HG002). Haplotype resolved assemblies for both genomes showed more than 99.99% base level accuracy with the reference genome. These results are comparable to the ones obtained with trio binning based methods such as TrioCanu.
We further applied our computational methods to sequencing data generated from Atlantic Bluefin Tuna (ABFT) genome. This genome is more heterozygous than human genome (0.01 vs 0.001). We obtained chromosome-scale assemblies for both the haplotypes with scaffold N50 of ~33.67 Mbp. Both the haplotypes when aligned to each other showed around 98.47% which is in concordance with the expected heterozygosity.
Uncover the role of SNPs, SVs, and Chromosome Phasing
Unsure how single nucleotide polymorphisms (SNPs), structural variants (SVs), or chromosome phasing impacts disease progression? Want to know if a SNP impacts a gene a megabase away? The Omni-C kit lets you dive deeper into the mechanisms of action to help you find answers. The improved coverage of Omni-C libraries enables genome-wide SNP calling, generate high-resolution data that can detect large structural variants, and deliver chromosome phasing information with low switch error rates – all with one library. Get a better understanding of the relationship between the genome and phenotype to potentially uncover the missing link for a particular disease. Elucidate mechanisms associated with SNPs that fall into non-coding regions to annotate their functional significance.
Omni-C Kit Overview
Dovetail Genomics has done all the protocol optimization and reagent qualification for you, so you can hit the ground running. The protocol eliminates the finicky sonication step used by other methods to improve reproducibility and is validated and well documented to ensure the data quality. Dovetail is dedicated to your success and has a team of support scientist ready for consultation whenever you need it.
Our Kits & Services
-
Description text goes here
-
Description text goes here